Cruz-Acuña Lab
Biomaterials and Tumorigenesis Lab
The Cruz-Acuña Lab focuses on integrating aspects of biomaterial engineering, cell and molecular biology, and 3D organoid biology to create engineered 3D organoid platforms that reveal novel mechanisms of tumorigenesis and organ development that can be leveraged for therapeutic studies and personalized medicine applications. Our primary focus is on the oral-esophageal tract, with potential applications to other gastrointestinal organs.
Our goal is to (1) help elucidate how cancer progresses within a dynamically evolving extracellular matrix to help inform the development of novel therapeutics, and (2) reveal novel mechanisms important for epithelial developmental patterning and organogenesis.
Our Lab is committed to building a diverse and inclusive work environment that achieves the goals of creating a scientific workforce that reflects all perspectives and experiences.
Research Areas
The Malignant Extracellular Matrix: Mechanisms to Translational Therapeutics
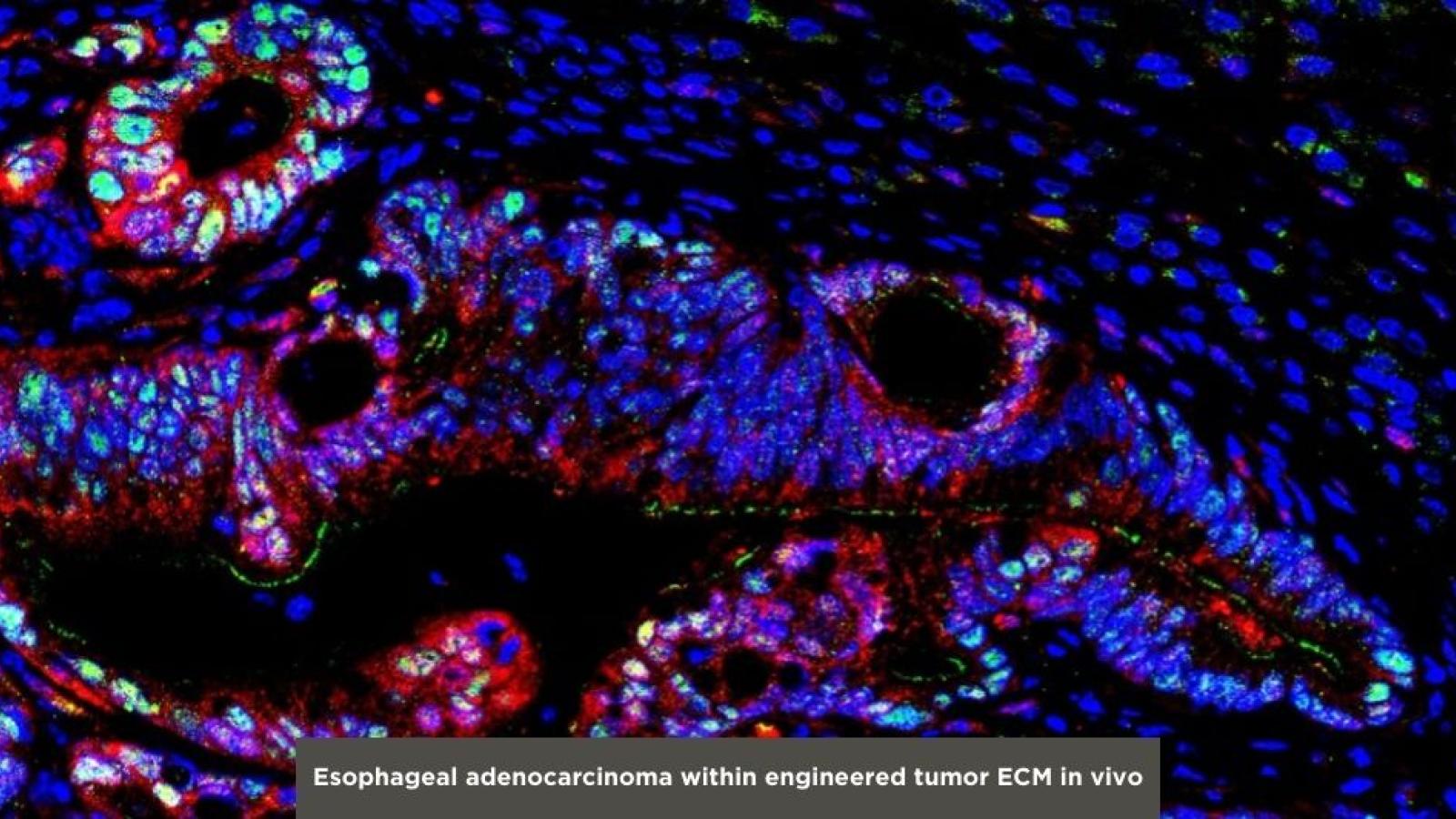
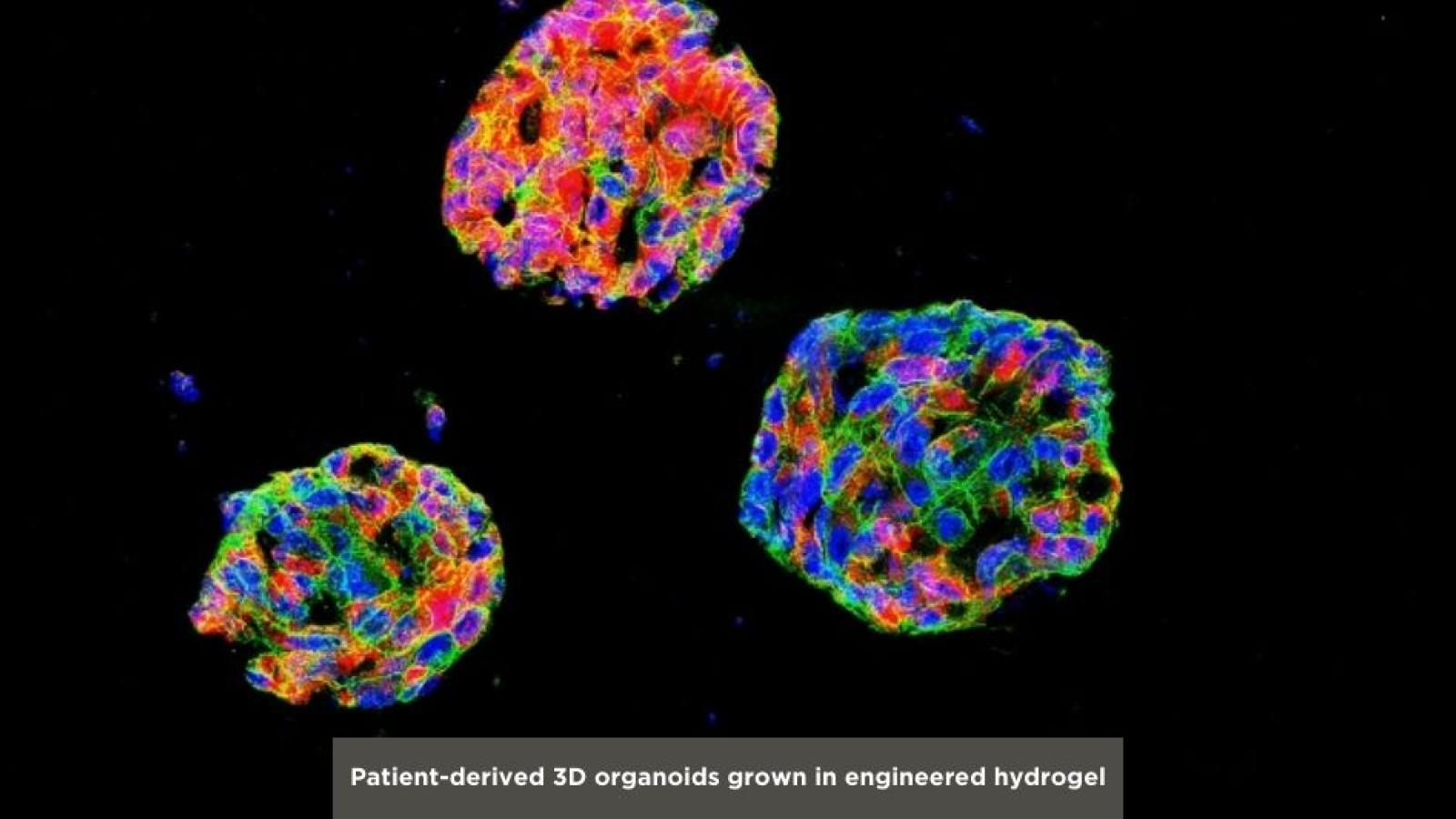
Recent studies have demonstrated that extracellular matrix (ECM) properties are associated with metaplasia-dysplasia-carcinoma sequence. Our work has revealed that increased ECM stiffness, a critical biomechanical property, modulates activation of oncogenes in esophageal cancer (Cruz-Acuña et al., JCI, 2023). This underscores a compelling need to understand mechanisms that foster metaplasia and dysplasia in the context of ECM properties. In this context, in our lab we utilize engineered hydrogels and patient-derived 3D organoids to elucidate how ECM properties contribute to pathophysiologic mechanisms that drive the metaplasia-dysplasia-carcinoma sequence in the oral-gastrointestinal tract. Moreover, our engineered 3D organoid culture system serves as a platform to explore the potential of matrix-activated genes as therapeutic targets in in vitro and in vivo settings. Importantly, the modular nature of the engineered biomaterial platform allows for potential adaption to the culture of 3D organoid models of other human diseases.
Genetic Engineering of hiPSCs to Study Cell Intrinsic Mechanisms of Metaplasia
Human induced pluripotent stem cells (hiPSCs) present a range of new opportunities for the study of metaplasia initiation in the context of cell intrinsic mechanisms. Their unlimited self-renewal and permissiveness for the introduction of point mutations (such as TP53 mutations) has provided a unique fashion to establish preneoplastic models that faithfully recapitulate mutation-specific cancer phenotypes. These findings are of particular importance as genomic analyses have found TP53 to be the most frequently mutated gene and an early event in intestinal metaplasia of the esophagus. Therefore, as studies suggest that ECM stiffening stabilizes mutant p53, we use hiPSC-derived esophageal organoids and esophageal organotypic raft culture systems to explore the cooperative role of ECM properties and TP53 mutations in tumorigenesis.
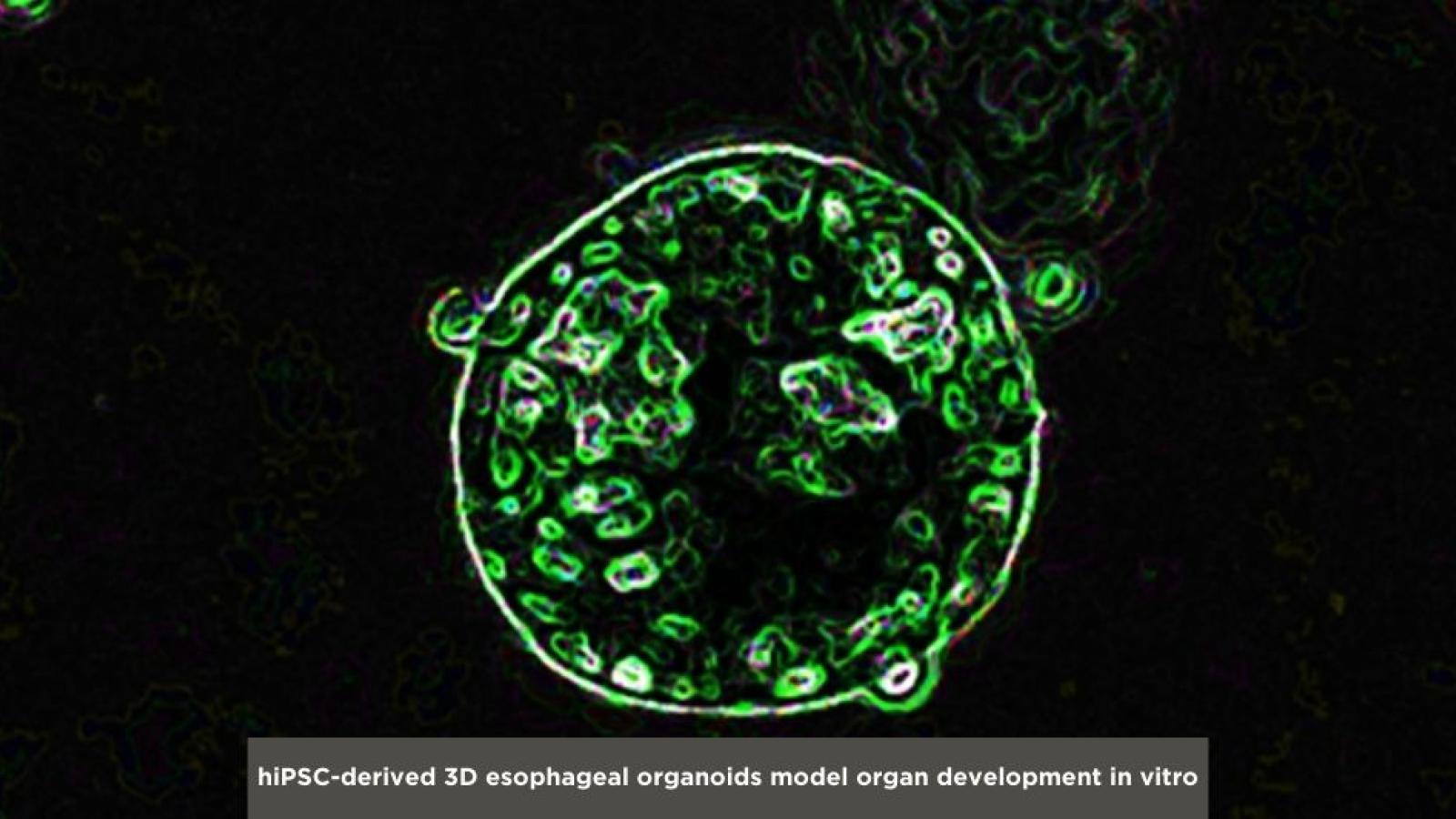
Elucidating Key Regulators of Early Organ Development
The human esophagus has a distinct tissue organization compared to rodent esophagus suggesting that developmental mechanisms may not be conserved between species. Therefore, little is known about the regulatory mechanisms important for patterning of the human foregut endoderm and later development of the early esophageal epithelium. In our lab, we use human pluripotent stem cells (hPSCs) as a powerful system to model early development of the human esophagus via sequential differentiation of hPSCs toward esophageal progenitor cells (EPCs). This system allows identification of key regulators of early development, differentiation, and keratinization of the human esophagus, with an emphasis on genes also involved in tumorigenesis.
Lab Members
Lab Members
Secunda W. Kariuki
- Staff Associate

Daniel Garcia
- Staff Associate

Select Publications
Cruz-Acuña, R., Kariuki, S.W., Sugiura, K., Karaiskos, S., Plaster, E.M., Loebel, C., Efe, G., Karakasheva, T.A., Gabre, J.T., Hu, J., Burdick, J.A., Rustgi, A.K. Engineered hydrogel reveals contribution of matrix mechanics to esophageal adenocarcinoma and identifies matrix-activated therapeutic targets. J Clin Invest, (2023) Oct 3:e168146. doi: 10.1172/JCI168146. Epub ahead of print. PMID: 37788109.
Kabir, M.F.*, Karami, A.L.*, Cruz-Acuña, R., Klochkova, A., Saxena, R., Mu, A., Murray, M.G., Cruz, J., Fuller, A.D., Clevenger, M.H., Chitrala, K.N., Tan, Y., Keith, K., Madzo, J., Huang, H., Jelinek, J., Karakasheva, T., Hamilton, K.E., Muir, A.B., Tétreault, M.P., Whelan, K.A. Single cell transcriptomic analysis reveals cellular diversity of murine esophageal epithelium. Nat Commun, (2022) Apr 20;13(1):2167. (*co-first authors)
Cruz-Acuña, R., Burdick, J.A., Vunjak-Novakovic, G. and Rustgi, A.K. Novel Technological Approaches Provide Insights on the Role of Extracellular Matrix Properties in Cancers and Therapeutics. iScience, (2021) May;24(5).
Cruz-Acuña, R., Mulero-Russe, A., Clark, A. Y., Zent, R. & García, A. J. Identification of matrix physicochemical properties required for renal epithelial cell tubulogenesis by using synthetic hydrogels. Journal of Cell Science, (2019) Oct 21;132(20).
Cruz-Acuña, R.*, Quirós, M.*, Huang, S., Siuda, V., Spence, J. R., Nusrat, A. and García, A. J. PEG-4MAL hydrogels for human organoid generation, culture, and in vivo delivery. Nature Protocols, (2018) Sep;13:2102-2119. (*co-first authors)
Cruz-Acuña, R.*, Quirós, M.*, Farkas, A. E., Dedhia, P. H., Huang, S., Siuda, D., García-Hernández, V., Spence, J. R., Nusrat, A. and García, A. J. Synthetic Hydrogels for Human Intestinal Organoid Generation and Colonic Wound Repair. Nature Cell Biology, (2017) Nov;19(11):1326-1335. (*co-first authors)